West Nile virus spread in Europe: Phylogeographic pattern analysis and key drivers
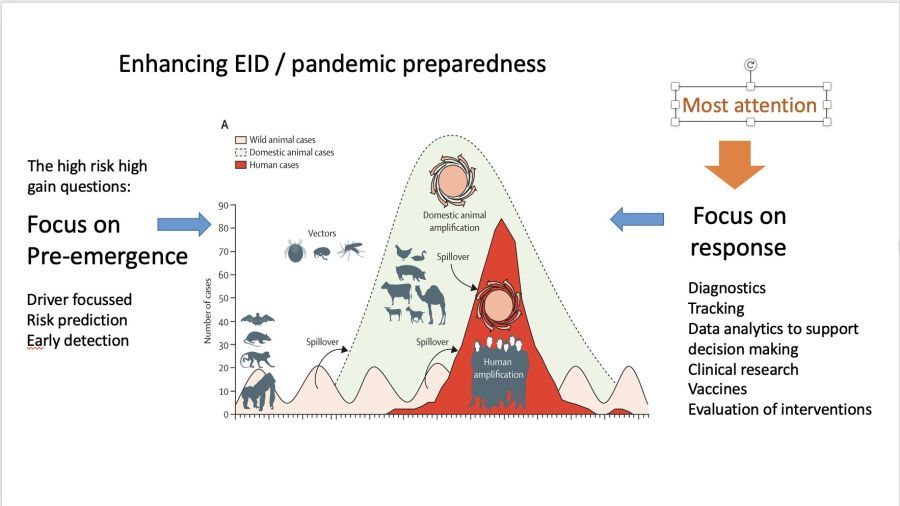
This publication is from a partner project of One Health PACT. Most epidemic preparedness focusses on enhancing response to outbreaks. In @EuropeVEO they try to figure out how to use combinations of data from multiple sources to get better at prediction outbreak risk, by focussing on understanding the ecology. VEO is coordinated by prof. Marion Koopmans: “In VEO we work through case studies of different types of emergence pathways: zoonotic wildlife borne, vector borne, potential emergence of pathogens from permafrost, and global expansion of a (novel) human pathogen through high density and highly connected urban areas. Each of these comes with a set of drivers for which we collect data that can help understand these pathways”.
Authors: Lu Lu 1 2, Feifei Zhang 2, Bas B Oude Munnink 3, Emmanuelle Munger 3, Reina S Sikkema 3, Styliani Pappa 4, Katerina Tsioka 4, Alessandro Sinigaglia 5, Emanuela Dal Molin 5, Barbara B Shih 1, Anne Günther 6, Anne Pohlmann 6, Ute Ziegler 7, Martin Beer 6, Rachel A Taylor 8, Frederic Bartumeus 9 10 11, Mark Woolhouse 2, Frank M Aarestrup 12, Luisa Barzon 5, Anna Papa 4, Samantha Lycett 1, Marion P.G. Koopmans
Abstract
Background: West Nile virus (WNV) outbreaks in birds, humans, and livestock have occurred in multiple areas in Europe and have had a significant impact on animal and human health. The patterns of emergence and spread of WNV in Europe are very different from those in the US and understanding these are important for guiding preparedness activities.
Methods: We mapped the evolution and spread history of WNV in Europe by incorporating viral genome sequences and epidemiological data into phylodynamic models. Spatially explicit phylogeographic models were developed to explore the possible contribution of different drivers to viral dispersal direction and velocity. A “skygrid-GLM” approach was used to identify how changes in environments would predict viral genetic diversity variations over time.
Findings: Among the six lineages found in Europe, WNV-2a (a sub-lineage of WNV-2) has been predominant (accounting for 73% of all sequences obtained in Europe that have been shared in the public domain) and has spread to at least 14 countries. In the past two decades, WNV-2a has evolved into two major co-circulating clusters, both originating from Central Europe, but with distinct dynamic history and transmission patterns. WNV-2a spreads at a high dispersal velocity (88km/yr-215 km/yr) which is correlated to bird movements. Notably, amongst multiple drivers that could affect the spread of WNV, factors related to land use were found to strongly influence the spread of WNV. Specifically, the intensity of agricultural activities (defined by factors related to crops and livestock production, such as coverage of cropland, pasture, cultivated and managed vegetation, livestock density) were positively associated with both spread direction and velocity. In addition, WNV spread direction was associated with high coverage of wetlands and migratory bird flyways.
Conclusion: Our results suggest that-in addition to ecological conditions favouring bird- and mosquito- presence-agricultural land use may be a significant driver of WNV emergence and spread. Our study also identified significant gaps in data and the need to strengthen virological surveillance in countries of Central Europe from where WNV outbreaks are likely seeded. Enhanced monitoring for early detection of further dispersal could be targeted to areas with high agricultural activities and habitats of migratory birds.
Read the whole publication here.